50s ribosomal subunit
Thank you for visiting nature.
The BipA B PI- i nducible p rotein A protein is highly conserved in a large variety of bacteria and belongs to the translational GTPases, based on sequential and structural similarities. Despite its conservation in bacteria, bipA is not essential for cell growth under normal growth conditions. Our recent studies revealed that BipA is a novel ribosome-associating GTPase, whose expression is cold-shock-inducible and involved in the incorporation of the ribosomal protein r-protein L6. However, the precise mechanism of BipA in 50S ribosomal subunit assembly is not completely understood. In this study, to demonstrate the role of BipA in the 50S ribosomal subunit and possibly to find an interplaying partner s , a genomic library was constructed and suppressor screening was conducted. Through screening, we found a suppressor gene, rplT , encoding r-protein L20, which is assembled at the early stage of ribosome assembly and negatively regulates its own expression at the translational level.
50s ribosomal subunit
The structures of ribosomal proteins and their interactions with RNA have been examined in the refined crystal structure of the Haloarcula marismortui large ribosomal subunit. The protein structures fall into six groups based on their topology. The 50S subunit proteins function primarily to stabilize inter-domain interactions that are necessary to maintain the subunit's structural integrity. An extraordinary variety of protein-RNA interactions is observed. Electrostatic interactions between numerous arginine and lysine residues, particularly those in tail extensions, and the phosphate groups of the RNA backbone mediate many protein-RNA contacts. Base recognition occurs via both the minor groove and widened major groove of RNA helices, as well as through hydrophobic binding pockets that capture bulged nucleotides and through insertion of amino acid residues into hydrophobic crevices in the RNA. Primary binding sites on contiguous RNA are identified for 20 of the 50S ribosomal proteins, which along with few large protein-protein interfaces, suggest the order of assembly for some proteins and that the protein extensions fold cooperatively with RNA. The structure supports the hypothesis of co-transcriptional assembly, centered around L24 in domain I. Finally, comparing the structures and locations of the 50S ribosomal proteins from H. These comparisons illustrate that identical RNA structures can be stabilized by unrelated proteins. Abstract The structures of ribosomal proteins and their interactions with RNA have been examined in the refined crystal structure of the Haloarcula marismortui large ribosomal subunit.
Timsit, Y. Another line of evidence for this is that deletion of rluC suppresses the growth defects of a bipA -deleted mutant at low temperature Krishnan and Flower,whose gene product is a pseudouridine synthase modifying nucleotides U, Y, and U in 23S rRNA Conrad et al, 50s ribosomal subunit.
Bacteria harbor a number GTPases that function in the assembly of the ribosome and are essential for growth. Homologs of this protein are also implicated in the assembly of the large subunit of the mitochondrial and eukaryotic ribosome. We present here the cryo-electron microscopy structure of RbgA bound to a Bacillus subtilis 50S subunit assembly intermediate 45S RbgA particle that accumulates in cells upon RbgA depletion. Binding of RbgA at the P site of the immature particle stabilizes functionally important rRNA helices in the A and P-sites, prior to the completion of the maturation process of the subunit. The structure also reveals the location of the highly conserved N-terminal end of RbgA containing the catalytic residue Histidine 9.
Federal government websites often end in. The site is secure. Ribosomes are among the largest and most dynamic molecular motors. The structure and dynamics of translation initiation and elongation are reviewed. Three ribosome motions have been identified for initiation and translocation. A reversible ratcheting motion was seen between the 30S and the 50S subunits that slide relative to one another. The 30S—50S intersubunit contacts regulate translocation. Divergence of class I and class II aaRS enzymes and coevolution of the genetic code are described by analysis of ancient archaeal species. We offer a general and conceptual review of translation in prokaryotic systems.
50s ribosomal subunit
It is the site of inhibition for antibiotics such as macrolides , chloramphenicol , clindamycin , and the pleuromutilins. Despite having the same sedimentation rate, bacterial and archaeal ribosomes can be quite different. X-ray crystallography has yielded electron density maps allowing the structure of the 50S in Haloarcula marismortui archaeon to be determined to 2.
Tech fleece shirt
Add comment Close comment form modal. Ban, N. PLos Genet. Stark, H. Thus, if the 45S particles are competent for maturation, the model predicts that all of the r-proteins incorporated into this particle will show increased P values in the RbgA-limiting condition. The max lab curves were calculated using Equation 2 :. Using the cryo-EM maps, we produced molecular models for class A and B assembly intermediates Supplemental Table S1 and used these models to produce temperature maps that measured how much the rRNA regions present in these structures deviated from the conformation of the mature 50S subunit Supplementary Figure S3. Suppression of delta bipA phenotypes in Escherichia coli by abolishment of pseudouridylation at specific sites on the 23S rRNA. Bridge B1a utilizes helix 38 and connects the CP to protein S13 in the head of the 30S subunit; bridge B2a, an essential interaction for 70S stability, connects helix 69 and the decoding site of the 30S subunit; and bridge B7a links helix 68 with helix 23 in the 30S subunit 26 , At this GTP concentration, the obtained values are within the linear range of the assay The fractions corresponding to the 45S particles in the gradient were collected, pooled and centrifuged for 16 h at g in a Beckman MLA rotor. While these structural changes are more subtle than those observed in the CP, these data indicates that the GAR is also malformed in this immature particle. The reference maps for the projection matching refinements were produced from the particles in each subclass using a maximum likelihood-based refinement approach 30 , 33 to prevent model bias. Amsterdam: North-Holland;
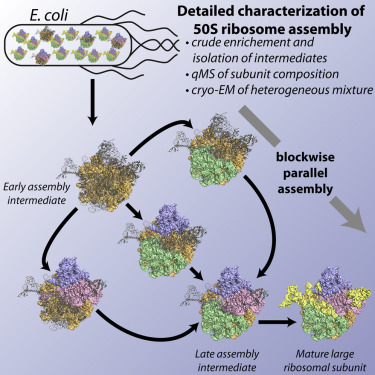
The assembly of ribosomal subunits is an essential prerequisite for protein biosynthesis in all domains of life. Although biochemical and biophysical approaches have advanced our understanding of ribosome assembly, our mechanistic comprehension of this process is still limited. Here, we perform an in vitro reconstitution of the Escherichia coli 50S ribosomal subunit.
We are also thankful to Richard Fekete for suggestions about the primer extension method. J-IO reviewed the results and manuscript. Similarly, helices 81, 84 and 85 located at the core of the CP adopted conformations distinct from the mature structure Figure 5 A; Supplementary Figure S8. Parmeggiani A. To determine the relationship between protein occupancy in the 45S and precursor pool size upon RbgA depletion, we plotted these quantities against one another Figure 2 E. D Struct. The acceptor stem of the E-site tRNA binds in a pocket formed by helices 68, 74, 75, 76 and 88 and the two r-proteins, L28 and L35 Figure 3. Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily. Despite its conservation in bacteria, bipA is not essential for cell growth under normal growth conditions.

The authoritative answer, curiously...