Allen brain atlas
Six years and 32 million cells later, scientists have created the first full cellular map of a mammalian brain.
Allen Brain Atlas has enormous potential to help unlock the mysteries of neurological diseases and disorders affecting millions worldwide. The Institute today announced the completion of the groundbreaking Allen Brain Atlas, a Webbased, three-dimensional map of gene expression in the mouse brain. Detailing more than 21, genes at the cellular level, the Atlas provides scientists with a level of data previously not available. About 26 percent of American adults — close to 58 million people — suffer from a diagnosable mental disorder in a given year. The project has already led to several significant new findings about the brain. It reveals that 80 percent of genes are turned on in the brain, much higher than the 60 to 70 percent scientists previously believed. It indicates that very few genes are turned on in only one region of the brain — paving the way for additional insight about the benefits and potential side effects of drug treatments.
Allen brain atlas
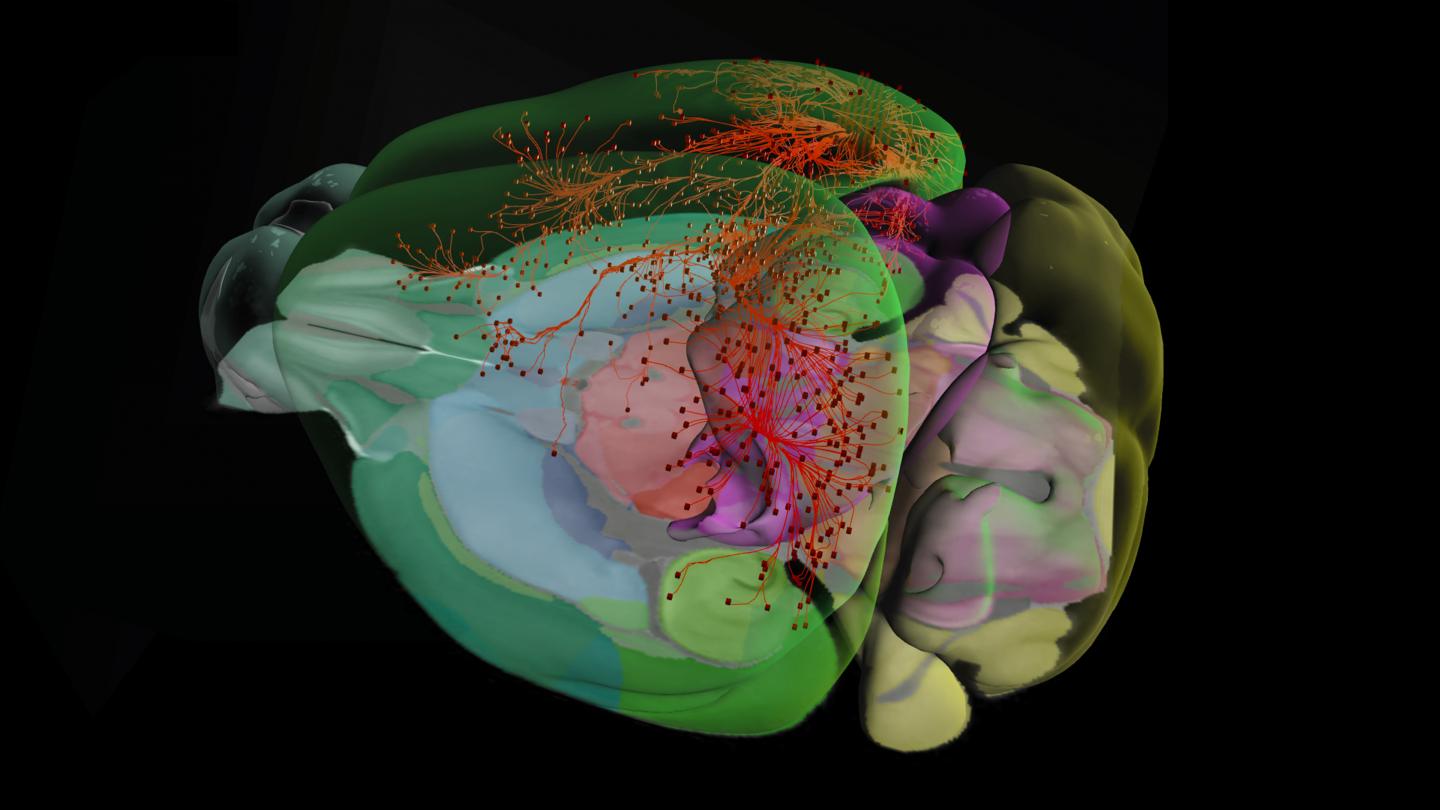
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. The mammalian brain consists of millions to billions of cells that are organized into many cell types with specific spatial distribution patterns and structural and functional properties 1 , 2 , 3. Here we report a comprehensive and high-resolution transcriptomic and spatial cell-type atlas for the whole adult mouse brain. The cell-type atlas was created by combining a single-cell RNA-sequencing scRNA-seq dataset of around 7 million cells profiled approximately 4. The atlas is hierarchically organized into 4 nested levels of classification: 34 classes, subclasses, 1, supertypes and 5, clusters. We systematically analysed the neuronal and non-neuronal cell types across the brain and identified a high degree of correspondence between transcriptomic identity and spatial specificity for each cell type. The results reveal unique features of cell-type organization in different brain regions—in particular, a dichotomy between the dorsal and ventral parts of the brain. The dorsal part contains relatively fewer yet highly divergent neuronal types, whereas the ventral part contains more numerous neuronal types that are more closely related to each other. Our study also uncovered extraordinary diversity and heterogeneity in neurotransmitter and neuropeptide expression and co-expression patterns in different cell types.
Even before its announced completion, the Atlas was receiving more than 4 million hits monthly and being accessed by approximately scientists on any given work day, allen brain atlas. Characterization of neural stem cells and their progeny in the sensory circumventricular organs of adult mouse.
The Allen mouse brain atlas is a comprehensive digital resource that provides detailed information on the structure and function of the mouse brain. The Allen Mouse Brain Common Coordinate Framework is the backbone of spatially-focused workflows and tools that support spatial registration of new data to the atlas framework, semi-automatic analyses using the brain region hierarchy and delineations, and visualisation of extracted data in 3D. The atlas is incorporated in the QuickNII tool for spatial registration of serial 2D images, and employed in the QUINT workflow for extracting and quantifying labelled objects from images registered to the atlas. Originally built as a backend service for the interactive atlas viewer siibra-explorer, the API has been documented for connecting the brain atlases to other applications and web services. Sign up now for complete access to our tools and services. All tools and software Mouse Brain Atlas Overview. Related tools The Allen Mouse Brain Common Coordinate Framework is the backbone of spatially-focused workflows and tools that support spatial registration of new data to the atlas framework, semi-automatic analyses using the brain region hierarchy and delineations, and visualisation of extracted data in 3D.
The Allen Mouse and Human Brain Atlases are projects within the Allen Institute for Brain Science which seek to combine genomics with neuroanatomy by creating gene expression maps for the mouse and human brain. Allen and the first atlas went public in September The atlases are free and available for public use online. In , Paul Allen gathered a group of scientists, including James Watson and Steven Pinker , to discuss the future of neuroscience and what could be done to enhance neuroscience research Jones During these meetings David Anderson from the California Institute of Technology proposed the idea that a three-dimensional atlas of gene expression in the mouse brain would be of great use to the neuroscience community. The project was set in motion in with a million dollar donation by Allen through the Allen Institute for Brain Science. The project used a technique for mapping gene expression developed by Gregor Eichele and colleagues at the Max Planck Institute for Biophysical Chemistry in Goettingen, Germany. The technique uses colorimetric in situ hybridization to map gene expression. The project set a 3-year goal of finishing the project and making it available to the public. An initial release of the first atlas, the mouse brain atlas, occurred in December
Allen brain atlas
A lightweight python module to interact with atlases for systems neuroscience. The brainglobe atlas API brainglobe-atlasapi provides a common interface for programmers to download and process brain atlas data from multiple sources. Full information can be found in the documentation.
Usps passport application walk in
The Allen Brain Atlas lets researchers view the areas of differing expression in the brain which enables the viewing of neural connections throughout the brain. To solve this problem, we leveraged the established cell-type hierarchy and performed imputation iteratively, first at top level and then within each class and subclass. Lein, E. Retrieved 20 April Except for the default gene set, the remaining genes were largely ordered with decreasing predictive power. In , Paul Allen gathered a group of scientists, including James Watson and Steven Pinker , to discuss the future of neuroscience and what could be done to enhance neuroscience research Jones We also developed a hierarchical version of this approach to assign cell-type identities for MERFISH, Multiome snRNA-seq, or any external datasets to the 10xv3 dataset as reference, using different gene lists based on the contexts. Kempermann, G. Other clusters , , and in the Astro-NT subclass are also localized at the pia with high expression of Gfap , which we hypothesize to be ILAs outside telencephalon. We also systematically identified all clusters that produce modulatory neurotransmitters Fig. The concentration of the resuspended cells was quantified, and cells were immediately loaded onto the 10x Genomics Chromium controller. Characterization of A11 neurons projecting to the spinal cord of mice.
The Allen Institute for Brain Science was established in with a goal to accelerate neuroscience research worldwide with the release of large-scale, publicly available atlases of the brain. Our research teams continue to conduct investigations into the inner workings of the brain to understand its components and how they come together to drive behavior and make us who we are. One of our core principles is Open Science: We publicly share all the data, products, and findings from our work.
Single-cell sequencing-based technologies will revolutionize whole-organism science. Cold Spring Harb. Class order was slightly altered to have less emphasize on neurotransmitter type and more on region specificity. Data and publications from the Allen Brain Observatory reveal new complexity of neuronal circuitry. Langfelder, P. The project set a 3-year goal of finishing the project and making it available to the public. We then attempted to select one DEG in each direction for any remaining pairs of clusters not covered by the selected genes using the same function. Mei, C. The atlas is also geared toward furthering research into mental health disorders and brain injuries such as Alzheimer's disease , autism, schizophrenia and drug addiction. Unveiled in July , the Allen Mouse Spinal Cord Atlas was the first genome-wide map of the mouse spinal cord ever constructed. For step 6, if one cluster had fewer than the minimal number of cells in a dataset 4 cells for 10xv3 and 10 cells for 10xv2 , then this dataset was not used for differentially expressed gene computation for all pairs involving the given cluster. Detailed cellular maps of the entire human brain reveal clues Molecular architecture of the mouse nervous system. We also identified tanycyte-like ependymal cell clusters that are specifically located in other CVOs Fig. Next, we used that same approach to assign each cell mapped to a non-anchor cluster to the region annotation dominating its immediate surrounding.

0 thoughts on “Allen brain atlas”