Allosteric
Federal government websites often end in.
These examples are programmatically compiled from various online sources to illustrate current usage of the word 'allosteric. Send us feedback about these examples. Accessed 4 Mar. Subscribe to America's largest dictionary and get thousands more definitions and advanced search—ad free! See Definitions and Examples ». Log In.
Allosteric
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Allostery in proteins influences various biological processes such as regulation of gene transcription and activities of enzymes and cell signaling. Computational approaches for analysis of allosteric coupling provide inexpensive opportunities to predict mutations and to design small-molecule agents to control protein function and cellular activity. We develop a computationally efficient network-based method, Ohm, to identify and characterize allosteric communication networks within proteins. Unlike previously developed simulation-based approaches, Ohm relies solely on the structure of the protein of interest. We use Ohm to map allosteric networks in a dataset composed of 20 proteins experimentally identified to be allosterically regulated. Our webserver, Ohm. David Ding, Ada Y. Shaw, … Debora S. If naturally selected sequences fold into unique structures, then both sequences and structures also possess information about folding dynamics, although this relationship remains an enigma.
This process is repeated 10 4 times. Dokholyan, allosteric, N. Hidden allosteric All articles with unsourced statements Articles with unsourced statements from April
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Allosteric communication between distant sites in proteins is central to biological regulation but still poorly characterized, limiting understanding, engineering and drug development 1 , 2 , 3 , 4 , 5 , 6. An important reason for this is the lack of methods to comprehensively quantify allostery in diverse proteins.
Several criteria must be met for a chemical reaction to happen. Obviously, the reactants must first find one another in space. Chemicals in solutions don't "plan" these collisions, they happen at random. The rate frequency of collisions per second at which two reactants find one another will depend on their velocity determined by temperature and their concentration. In this Biology course, we'll assume temperature is a constant. Secondly, in addition to colliding, the molecules probably have to collide at the correct orientations, as not all collisions are potentially productive. Thirdly, the molecules have to have sufficient energy to form the transition state. If the transition state is significantly above the average energy of the molecules which will be fairly uniform- a narrow distribution very very few of the molecules will have sufficient energy to form the unstable, high tension, distorted "transition state". Thus, even if the reactants collide frequently, and the reaction is energetically favorable, a reaction with a activation energy significantly above the average energy of the reactants is not going to progress on a timescale suitable for the life of a cell. This is actually, and perhaps surprisingly, good news for the cell.
Allosteric
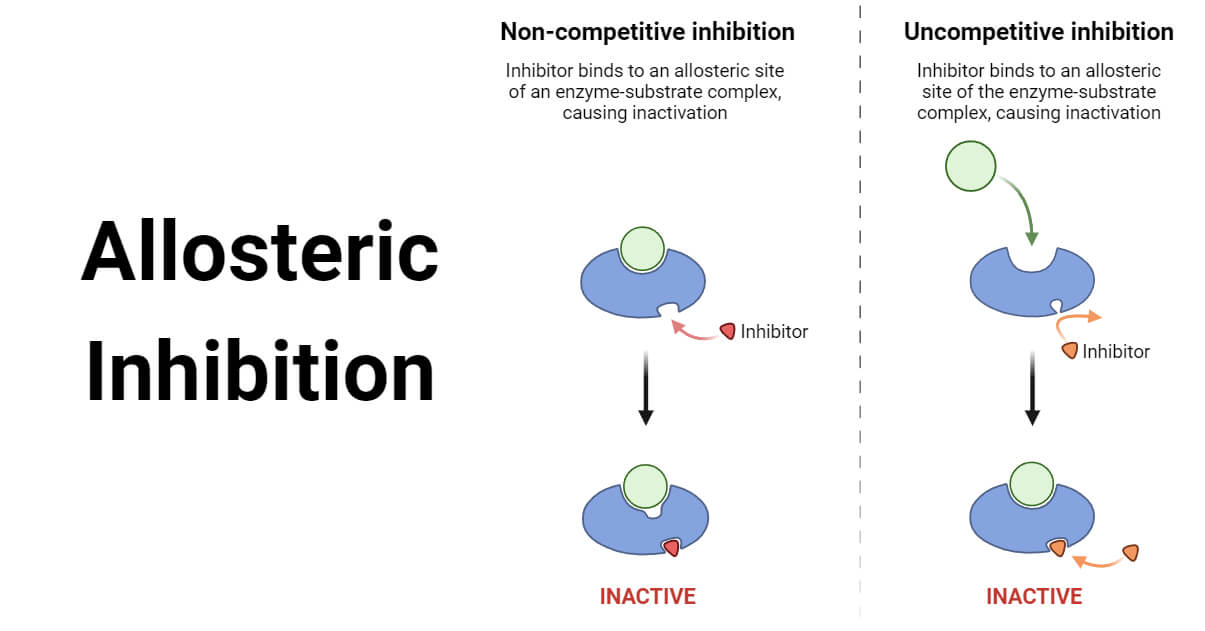
In pharmacology and biochemistry , allosteric modulators are a group of substances that bind to a receptor to change that receptor's response to stimuli. Some of them, like benzodiazepines or alcoholic beverages , function as psychoactive drugs. Modulators and agonists can both be called receptor ligands. Allosteric modulators can be 1 of 3 types either: positive, negative or neutral. Positive types increase the response of the receptor by increasing the probability that an agonist will bind to a receptor i. Neutral types don't affect agonist activity but can stop other modulators from binding to an allosteric site. Some modulators also work as allosteric agonists and yield an agonistic effect by themselves.
Ariat australia
Rational design of a ligand-controlled protein conformational switch. Another instance in which negative allosteric modulation can be seen is between ATP and the enzyme phosphofructokinase within the negative feedback loop that regulates glycolysis. Science Bandaru, P. Natl Acad. A non-regulatory allosteric site is any non-regulatory component of an enzyme or any protein , that is not itself an amino acid. Biochemical Society Transactions. Analysis of activated mutants by 19F NMR and protein engineering. However the molecular basis for conversion between the two states is not well understood. There are 6 prominent peaks in ACI values along the sequence Fig. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Morzan Victor S. Protein context shapes the specificity of SH3 domain-mediated interactions in vivo.
These examples are programmatically compiled from various online sources to illustrate current usage of the word 'allosteric. Send us feedback about these examples.
Starting from residue n , the propagation process is performed, and the current path is added to S if the end of the path is m. The dataset includes seven monomers, two dimers, one trimer, seven tetramers, two hexamers, and one dodecamer proteins. The origin of protein interactions and allostery in colocalization. Further information on research design is available in the Nature Research Reporting Summary linked to this article. Abstract Allosteric communication between distant sites in proteins is central to biological regulation but still poorly characterized, limiting understanding, engineering and drug development 1 , 2 , 3 , 4 , 5 , 6. A stack S is constructed to store all the pathways that pass through the active site and the allosteric site. After tar archives have been extracted, three files are obtained: a structure file for the query protein, site information for the predicted results, and a. European Biophysics Journal. First, contacts are extracted from the tertiary structure of the protein. Comments By submitting a comment you agree to abide by our Terms and Community Guidelines. This is in reference to the fact that the regulatory site of an allosteric protein is physically distinct from its active site. Signal transduction by allosteric receptor oligomerization. Peer Review File. Ligands are shown as black sticks.

0 thoughts on “Allosteric”