Clin var
Genome Medicine volume 15Article number: 51 Cite this article. Metrics details.
Federal government websites often end in. The site is secure. ClinVar accessions submissions reporting human variation, interpretations of the relationship of that variation to human health and the evidence supporting each interpretation. The database is tightly coupled with dbSNP and dbVar, which maintain information about the location of variation on human assemblies. Each ClinVar record represents the submitter, the variation and the phenotype, i. The submitter can update the submission at any time, in which case a new version is assigned. Interactive and programmatic access to the interpretation of medically important human variation is critical to realizing the promise of genomic medicine.
Clin var
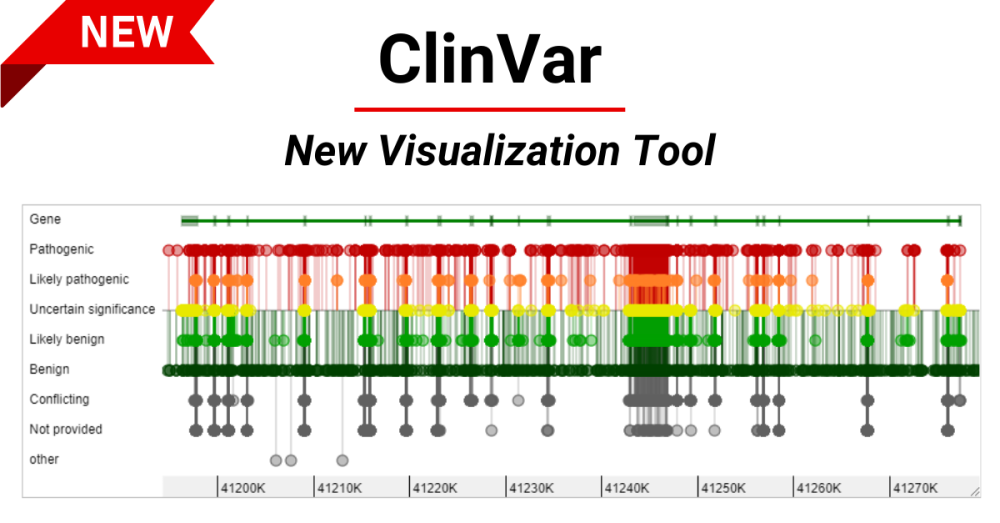
Interpretations of the clinical significance of variants are submitted by clinical testing laboratories, research laboratories, expert panels and other groups. ClinVar aggregates data by variant-disease pairs, and by variant or set of variants. Data aggregated by variant are accessible on the website, in an improved set of variant call format files and as a new comprehensive XML report. ClinVar recently started accepting submissions that are focused primarily on providing phenotypic information for individuals who have had genetic testing. ClinVar continues to make improvements to its search and retrieval functions. Several new fields are now indexed for more precise searching, and filters allow the user to narrow down a large set of search results. ClinVar 1 , 2 is a freely available, public archive of human genetic variants and interpretations of their significance to disease. Assertions of the clinical significance of a variant or set of variants are submitted to ClinVar by clinical testing laboratories, research laboratories, locus-specific databases, expert panels and other groups. Submissions include a description of the variant s ; the condition for which the variant was interpreted; the interpretation of the clinical significance of the variant, with the option to provide mode of inheritance; and evidence for that interpretation. ClinVar aggregates submissions based both on the variant and the variant-condition pair, and calculates an aggregate interpretation to indicate whether there is consensus or disagreement among submitters for an interpretation. Review status is based on submission of the criteria used by the submitter to classify variants, consensus across submitters in the interpretation of the variant and whether an expert panel or practice guideline-providing group has interpreted the variant.
However, because the 1KGP hemizygous South Asian ancestry male NA has not yet reached the maximal age of onset, a pathogenic classification cannot be ruled clin var.
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Automated variant filtering is an essential part of diagnostic genome-wide sequencing but may generate false negative results. We sought to investigate whether some previously identified pathogenic variants may be being routinely excluded by standard variant filtering pipelines.
Federal government websites often end in. Before sharing sensitive information, make sure you're on a federal government site. The site is secure. Go to the search box in the gray area at the top of the page. Just type your search term and click on the Search button to the right of the search box. ClinVar can be searched with terms like:. In other words, when you enter a term or phrase of interest in the query box, that term or phrase will be processed to retrieve records that contain or have some relationship to the word s you entered. The information is also organized into information categories or fields, so that queries can be constructed that retrieve records only if the term of interest occurs in that field. If you know the name of the field, you can enter that field name yourself. Otherwise you can use the Advanced page to help you build your query.
Clin var
Federal government websites often end in. Before sharing sensitive information, make sure you're on a federal government site. The site is secure. ClinVar is a freely accessible, public archive of reports of human variations classified for diseases and drug responses, with supporting evidence. ClinVar thus facilitates access to and communication about the relationships asserted between human variation and observed conditions, and the history of those assertions. ClinVar processes submissions reporting variants found in patient samples, classifications for diseases and drug responses, information about the submitter, and other supporting data. The variants described in submissions are mapped to reference sequences, and reported according to the HGVS standard. ClinVar presents the data on the website for interactive users, and on the FTP site and by API for those wishing to use ClinVar programmatically in daily workflows and other local applications. ClinVar works in collaboration with interested organizations to meet the needs of the medical genetics community as efficiently and effectively as possible.
The midnight club ekşi
S10A, B. The database is tightly coupled with dbSNP and dbVar, which maintain information about the location of variation on human assemblies. We also considered whether P or LP variants led to a larger number of indicated affected individuals. Since then, a growing number of laboratories have adopted these guidelines [ 9 ]. Additional file 2: Table S2. As has been well documented previously, erroneous classifications of pathogenicity exist in most variant databases 17 due to insufficient evidence being available at the time of classification. We observed that African ancestry individuals have a significantly increased chance of being incorrectly indicated to be affected by a screened IEM when HGMD variants are used. Fortunately, these misclassified variants were eventually corrected. Advance article alerts. HGMD variants that were removed from the database were classified as R. Variant databases are under continuous development and growth [ 22 , 23 ]. Am J Hum Genet. Each ClinVar record represents the submitter, the variation and the phenotype, i.
.
Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. ANA is currently an employee of Illumina, Inc. Figure 1. We identified more individuals than expected compared to the incidence of screened IEMs, which allowed us to assess the specificity of each database. Results Of these ClinVar pathogenic variants, 3. Copyright Published by Oxford University Press Several new fields are now indexed for more precise searching, and filters allow the user to narrow down a large set of search results. Filters are applied per Variation ID, not per submission. Note that allele origin refers to an observation of a variant, not the variant itself, so the same variant may have been reported both as germline and as somatic. These gaps impede the identification of benign variants from their high allele frequency in these populations. Petrovski S, Goldstein DB. Variant databases have taken different approaches to address misclassifications.

You have thought up such matchless phrase?
Many thanks for the information, now I will know.