Conserved domain database
Conserved Domains and Protein Classification. HOW TO. Citing the Resources.
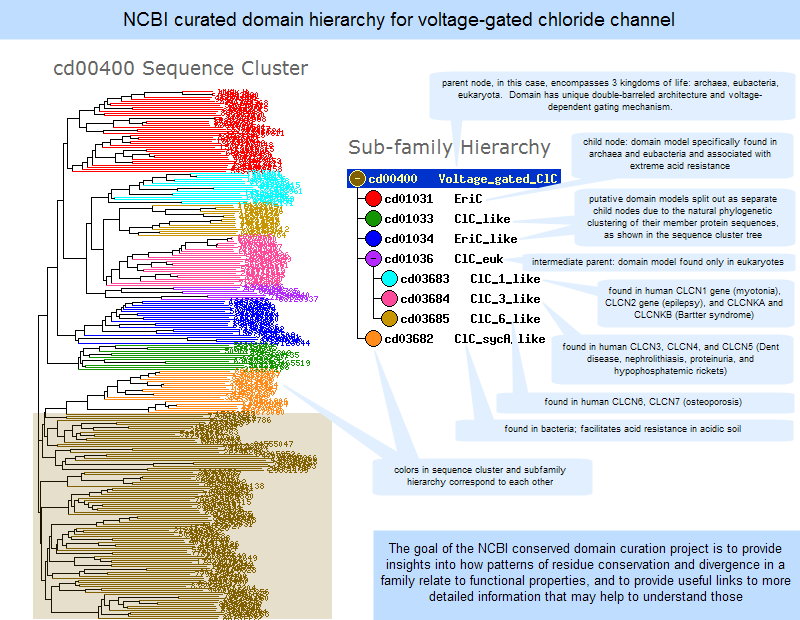
The Conserved Domain Database CDD is a database of well-annotated multiple sequence alignment models and derived database search models, for ancient domains and full-length proteins. These two classifications coincide rather often, as a matter of fact, and what is found as an independently folding unit of a polypeptide chain also carries specific function. Domains are often identified as recurring sequence or structure units, which may exist in various contexts. In molecular evolution such domains may have been utilized as building blocks, and may have been recombined in different arrangements to modulate protein function. CDD defines conserved domains as recurring units in molecular evolution, the extents of which can be determined by sequence and structure analysis. Manually curated models are organized hierarchically if they describe domain families that are clearly related by common descent.
Conserved domain database
CDD has been available publicly for over 20 years and has grown substantially during that time. Maintaining an archive of pre-computed annotation continues to be a challenge and has slowed down the cadence of CDD releases. CDD aims to collect a comprehensive set of protein and domain family models, and it does allow for considerable redundancy in the model set, to ensure good coverage of the protein space. Models that provide significantly overlapping annotation are clustered into protein domain superfamilies, and when domain annotation fails to exceed critical model-specific score thresholds, CDD by default reports superfamily annotation rather than individual model hits. For each model, we compute a consensus sequence, which is used for display purposes only, and reflects the length of the position-specific score matrix PSSM. While consensus sequences are visible and made available, CDD is not a sequence collection, but is rather meant to enrich the annotation of existing sequence collections. The current CDD version, v3. For CDD v3. The upcoming CDD release v3. Table 1 details the composition of CDD release v3. The sizes of these source databases vary considerably, as they were assembled for distinct purposes, and may only consider subsets of the protein space, such as limited by taxonomy. Neither of them is a complete accounting of protein and protein domain families, and neither is the aggregate of many resources, as 1 available protein sequence collections are expanding rapidly and becoming more diverse and 2 limited curation resources impact further growth. Table 2 shows the 20 largest classifications for common and functionally diverse domain families that have recently been updated or added to CDD.
Submit Cancel.
Toggle navigation. Repository details Conserved Domain database. General Institutions Terms Standards Name of repository. Additional name s. Repository URL. Subject s. The Conserved Domain Database is a resource for the annotation of functional units in proteins.
This page provides quick start guides for some common types of searches. The CDD Help document provides detailed descriptions of the database content, search system, and display formats. Once records of interest are retrieved, follow Entrez's "Links" to discover associations among previously disparate data. Conserved Domains and Protein Classification. HOW TO. How to use CDD: examples. Identify the putative function of a protein sequence.
Conserved domain database
A domain architecture is defined as the sequential order of conserved domains in a protein sequence. Regardless of which method you use, the results will display a list of similar domain architectures , which are ranked by the number of domains they share in common with the query protein's domain architecture. The results display also provides links to the proteins that have a each architecture. Click on any frame of the image below to link to corresponding sections in the CDART help document, which provide additional details about the input options and output display. Click on any frame of the image above to link to subsequent sections in this help document, which provide additional details about the input options and output display. Conserved Domains and Protein Classification.
Tunisia weather in december
Total Views 17, View a query protein sequence embedded within the multiple sequence alignment of a domain model. Lanczycki, Fu Lu, Gabriele H. Structural motifs are shown as double-headed arrows. The transporter classification database TCDB : update. The majority of the models included in the CDD collection do not require a very low word-score threshold in order to be identified as matches by members of the respective protein domain family. RefSeq: expanding the prokaryotic genome annotation pipeline reach with protein family model curation. Figure 1. Conserved Domains and Protein Classification. BMC Bioinformatics Jun 22;13 1 We maintain a live search system as well as an archive of pre-computed domain annotation for sequences tracked in NCBI's Entrez protein database, which can be retrieved for single sequences or in bulk. The conserved domain database in Permissions Icon Permissions.
The Conserved Domain Database CDD is a database of well-annotated multiple sequence alignment models and derived database search models, for ancient domains and full-length proteins.
CD-Search: protein domain annotations on the fly. To provide a non-redundant view of the data, CDD clusters similar domain models from various sources into superfamilies. Dachuan Zhang. HOW TO. Conserved Domains and Protein Classification. What is a conserved domain? Search ADS. Enhanced Publication. Conserved domain models utilized in CDD's annotation procedures may not yield significant scores with all sequence fragments that are true members of the respective family. While consensus sequences are visible and made available, CDD is not a sequence collection, but is rather meant to enrich the annotation of existing sequence collections. Pfam: the protein families database in To the extent possible under law, re3data. Epub Nov

In it something is. Thanks for the help in this question, the easier, the better �
Willingly I accept. The question is interesting, I too will take part in discussion. Together we can come to a right answer. I am assured.
I know, how it is necessary to act, write in personal