Neb double digest
Learn more. We are excited to announce that all reaction buffers are now BSA-free.
Ok this is going to be a long post so sit yourself down and prepare to be boggled by this mystery. I am currently trying to insert a 2. The pcDNA 3. I cut out the band using a new razor blade cutting the gel right on the 5. I get a nice clear product only at 5.
Neb double digest
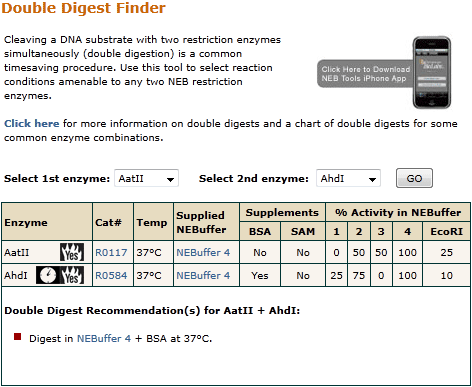
Digesting a DNA substrate with two restriction enzymes simultaneously double digestion is a common timesaving procedure. If you are using an enzyme that is not supplied with rCutSmart the Performance Chart for Restriction Enzymes rates the percentage activity of each restriction endonuclease in the four standard NEBuffers. Otherwise, choose an NEBuffer that results in the most activity for both enzymes. Set up reaction according to recommended protocol. If two different incubation temperatures are necessary, choose the optimal reaction buffer and set up reaction accordingly. Add the first enzyme and incubate at the desired temperature. If the enzyme is heat inactivatable, a heat inactivation step is recommended. Add the second enzyme and incubate at the recommended temperature. In most cases, DpnII requires a sequential digest. Set up a reaction using the restriction endonuclease that has the lowest salt concentration in its recommended buffer and incubate to completion. Adjust the salt concentration of the reaction using a small volume of a concentrated salt solution to approximate the reaction conditions of the second restriction endonuclease. Add the second enzyme and incubate to complete the second reaction. Do you want to sign out? Are you sure you want to delete this item?
The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. Drive your projects to completion faster with these high-fidelity fragments, neb double digest. I then proceed to do a negative control for my vector, so I ligate my vector by itself.
Through this new partnership we are pleased to offer you comprehensive next generation sequencing solutions. Unsure of what products are available? Confidently detect more with Archer NGS assay solutions for your solid tumor, blood cancer, immune profiling, and genetic disease research. Drive your projects to completion faster with these high-fidelity fragments. Shipping time is dependent on length and complexity of the dsDNA fragment s ordered.
Web pricing is applicable only to orders placed online at www. Most of our enzymes are supplied with one of four standard NEBuffers. Since rCutSmart Buffer includes Recombinant Albumin, there are also fewer tubes and pipetting steps to worry about. An exception occurred during the operation, making the result invalid. Check InnerException for exception details. Specifications and individual lot data from the tests that are performed for this particular product can be found and downloaded on the Product Specification Sheet, Certificate of Analysis, data card or product manual. Further information regarding NEB product quality can be found here. Specifications The Specification sheet is a document that includes the storage temperature, shelf life and the specifications designated for the product. Certificate of Analysis The Certificate of Analysis COA is a signed document that includes the storage temperature, expiration date and quality controls for an individual lot. The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed.
Neb double digest
Two restriction enzymes are used simultaneously to digest DNA in a single reaction. If your DNA concentration is too low you can increase the reaction volume to ul. Mix well by pipetting slowly up and down approximately 5x. Be gentle, and do not vortex. Spin the samples for 5 seconds in a balanced microcentrifuge, or flick them to collect the mixture at the bottom of the tube. Incubate at 37 degrees for at least 1 hour. For 3A assembly it is important you heat inactivate your samples after digestion.
Waitrose cellar
Use this tool when designing PCR reaction protocols to help determine the optimal annealing temperature for your amplicon. Both gBlocks and gBlocks HiFi Gene Fragments demonstrate consistent high sequence fidelity and purity across various lengths. This lead me to believe that perhaps my RE were not working properly so I used RE's from another lab and got the same result. Placing a K mixed base in the third position of codons eliminates the TAA and TGA stop codons from being included in the gene fragments libraries, leaving only TAG as the possible stop codon. NNK codons are commonly used in screens for codon substitutions to reduce the presence of some codon-rich amino acids, thereby reducing their overrepresentation in a particular library. Confidently detect more with Archer NGS assay solutions for your solid tumor, blood cancer, immune profiling, and genetic disease research. Hopefully you will see linearized vector and some uncut vector for one enzyme. The additional PCR step is not required to generate additional material and adds an additional enzymatic step to the protocol that can introduce errors. Web tools. In addition, supercoiled DNA may have varying rates of cleavage. Tm Calculator.
We have numerous interactive online tools for these and other questions in your daily lab work. Competitor Cross-Reference Tools. Use this tool to find the right products and protocols for each step digestion, end modification, ligation and transformation of your next traditional cloning experiment.
Then just cut out the linearized vector, gel purify, cut with the second enzyme and gel purify again. We do not recommend using PCR to add the overhangs, because PCR can preferentially amplify smaller byproducts in the prep, leading to issues with the cloning reaction and increasing the number of colonies you need to screen to find the correct full-length insert. What should I do if the IDT online ordering tool says that my sequence is too complex to order, but I need it as is? Up to 18 N or K mixed bases. The Harmonized Screening Protocol describes the gene sequence and customer screening practices that IGSC member companies employ to prevent the misuse of synthetic genes. Please see our Confidentiality Statement for more information. View FAQ. I then proceed to do a negative control for my vector, so I ligate my vector by itself. All gBlocks gene fragments FAQs. Product data High fidelity and purity for gene assembly Both gBlocks and gBlocks HiFi Gene Fragments demonstrate consistent high sequence fidelity and purity across various lengths. Fortunately, the large majority of sequences should be correct, so by sequencing a few more colonies you should find a correct clone.

Just that is necessary. An interesting theme, I will participate.