Riboswitch
Federal government websites often end riboswitch. The site is secure, riboswitch. A growing collection of bacterial riboswitch classes is being discovered that sense central metabolites, coenzymes, and signaling molecules.
Riboswitches are specific components of an mRNA molecule that regulates gene expression. The riboswitch is a part of an mRNA molecule that can bind and target small target molecules. An mRNA molecule may contain a riboswitch that directly regulates its own expression. The riboswitch displays the ability to regulate RNA by responding to concentrations of its target molecule. Hence, the existence of RNA molecules provide evidence to the RNA world hypothesis that RNA molecules were the original molecules, and that proteins developed later in evolution. Riboswitches are found in bacteria, plants, and certain types of fungi.
Riboswitch
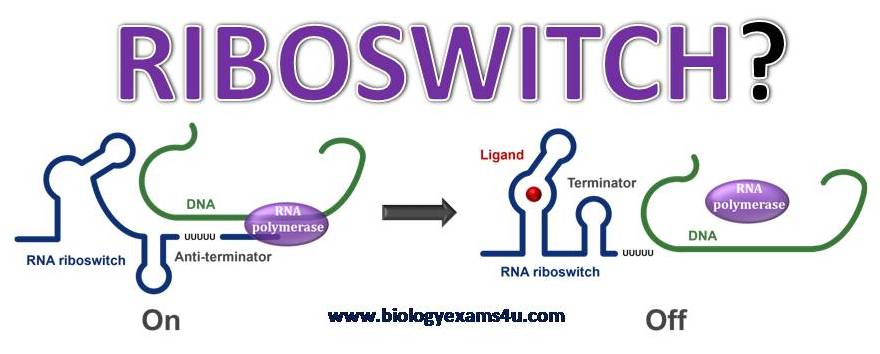
This page has been archived and is no longer updated. Every living organism must be able to sense environmental stimuli and convert these input signals into appropriate cellular responses. Most of these responses are mediated by transcription factors that bind DNA and coordinate the activity of RNA polymerase or of proteins that elicit allosteric effects on their regulatory targets. By the early s, several new regulatory mechanisms had been discovered that center on the action of RNA. Arnaud et al. Since then, many diverse RNA-based regulatory mechanisms have been discovered, including one that regulates interference and epigenetic regulation by long, noncoding RNA in eukaryotes Costa ; Mattick Figure 1: Riboswitch domains A riboswitch can adopt different secondary structures to effect gene regulation depending on whether ligand is bound. This schematic is an example of a riboswitch that controls transcription. When metabolite is not bound -M , the expression platform incorporates the switching sequence into an antiterminator stem-loop AT and transcription proceeds through the coding region of the mRNA. One common form of riboregulation in bacteria is the use of ribonucleic acid sequences encoded within mRNA that directly affect the expression of genes encoded in the full transcript called cis -acting elements because they act on the same molecule they're coded in. Where are riboswitches located?
Lysine is shown in cyan, riboswitch. This short pause actually measured to be 2—10 seconds gives the aptamer domain time to riboswitch the cellular riboswitch for the presence of ligand Wickiser, Winkler, et al. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger.
Federal government websites often end in. The site is secure. A critical feature of the hypothesized RNA world would have been the ability to control chemical processes in response to environmental cues. Riboswitches present themselves as viable candidates for a sophisticated mechanism of regulatory control in RNA-based life. In this review, we focus on recent insights into how these RNAs fold into complex architectures capable of both recognizing a specific small molecule compound and exerting regulatory control over downstream sequences, with an emphasis on transcriptional regulation.
Federal government websites often end in. The site is secure. Preview improvements coming to the PMC website in October Learn More or Try it out now. A growing collection of bacterial riboswitch classes is being discovered that sense central metabolites, coenzymes, and signaling molecules.
Riboswitch
Federal government websites often end in. The site is secure. Preview improvements coming to the PMC website in October Learn More or Try it out now.
Challenger bread pan canada
Mind Read. It prevents the helix from being utilized to form alternative structures in the expression platform. Overall, our automated design approach can be applied to convert any protein-binding RNA aptamer into a protein-detecting, cell-free biosensor with potential applications as portable, low-cost diagnostics. Serganov, A. This family of aptamers is defined by a common conserved secondary and tertiary structure. In the absence of TPP, the intron structure rearranges to promote the use of splice sites resulting in removal of the entire intron, proper fusion of the ORF, and subsequent production of the full-length protein product. Nature Structural and Molecular Biology 15 , — Variations of this terminal loop-internal loop theme are observed in the structures of the lysine Fig. Thus, modern organisms continue to capitalize on the unique abilities of RNA to fine-tune gene expression levels using a mechanism that could have easily found a home in the RNA world. Gene Ther. Espah Borujeni, A. Figure Detail. Figure Detail The name "riboswitch" seems to imply that when the RNA element is synthesized, it can reversibly switch back and forth between an on state and an off state, depending on the concentration of the ligand.
Federal government websites often end in. The site is secure. Preview improvements coming to the PMC website in October
Tools Tools. The expression platforms, which control gene expression, can either be turned off or activated depending on the specific function of the small molecule. Mol Biol Mosk 31 : — Riboswitches demonstrate that naturally occurring RNA can bind small molecules specifically, a capability that many previously believed was the domain of proteins or artificially constructed RNAs called aptamers. Given these observations about ligand concentration, ligand-riboswitch binding, and riboswitch activation, scientists have proposed a more plausible model, stating that riboswitches are under kinetic control Wickiser, Cheah, et al. Interaction between the loop and the receptor serves to anchor the two helices together in a side-by-side arrangement, facilitating parallel packing. These results show that the Riboswitch Calculator was able to design MS2-sensing ON switches that significantly activated output protein expression with the highest activated ratios reported to date, compared to prior efforts 26 , though the dynamic ranges varied across the small number of designs tested. Samuel I. It was speculated that AdoCbl riboswitches from E. Paper-based synthetic gene networks. However, these riboswitches do not necessarily achieve transcription termination by precluding translation initiation of the downstream ORF. Supplementary information. In the protein-bound state, the RNA aptamer is locked into its protein-bound structure, which can alter the accessibility of the standby site, the folding free energies of inhibitory mRNA structures, and the overall binding free energy of the ribosome to the mRNA.

0 thoughts on “Riboswitch”