Uniprot
The aim of the UniProt Knowledgebase is to provide users with a comprehensive, high-quality and freely accessible set of protein sequences annotated with functional information. In this article, uniprot, we describe significant updates uniprot we have made over uniprot last two years to the resource.
Hide the news. Posted Invalid Date -. Explore high-quality biological data resources. Evolution biology. Population genetics. Drug design. Medicinal chemistry.
Uniprot
Federal government websites often end in. The site is secure. UniProt releases are published every eight weeks. We provide customizable views and downloads in a range of formats via the website, and file sets at the FTP site www. The aim of the UniProt Knowledgebase is to provide users with a comprehensive, high-quality and freely accessible set of protein sequences annotated with functional information. In this publication we describe enhancements made to our data processing pipeline and to our website to adapt to an ever-increasing information content. The number of sequences in UniProtKB has risen to over million and we are working towards including a reference proteome for each taxonomic group. We continue to extract detailed annotations from the literature to update or create reviewed entries, while unreviewed entries are supplemented with annotations provided by automated systems using a variety of machine-learning techniques. In addition, the scientific community continues their contributions of publications and annotations to UniProt entries of their interest. The UniProt databases enable the research community to explore the diversity of life as described by the complement of proteins expressed by each organism. The UniRef databases cluster sequence sets at various levels of sequence identity and the UniProt Archive UniParc delivers a complete set of known unique sequences, including historical obsolete sequences. Data from selected resources are additionally integrated into UniProtKB records to add biological knowledge and associated metadata enabling the database to act as a central hub from which users can link out to other resources. Community functional annotation adds further value to the entry annotations. The integration of these data and the manual curation of protein features, such as functional domains and active sites, amino acid variants, ligand binding sites and post-translational modifications PTMs in the UniProt record, provide our users with mechanistic insights into how, for example, specific variants can lead to disease or resistance to a drug or to a pathogen. In , structural predictions were added from AlphaFold, a machine-learning system developed by DeepMind that predicts a protein's 3-dimensional 3D structure from its amino acid sequence 1.
Steinegger M, uniprot. Please check for further notifications by email. Natale, Karen Ross, C.
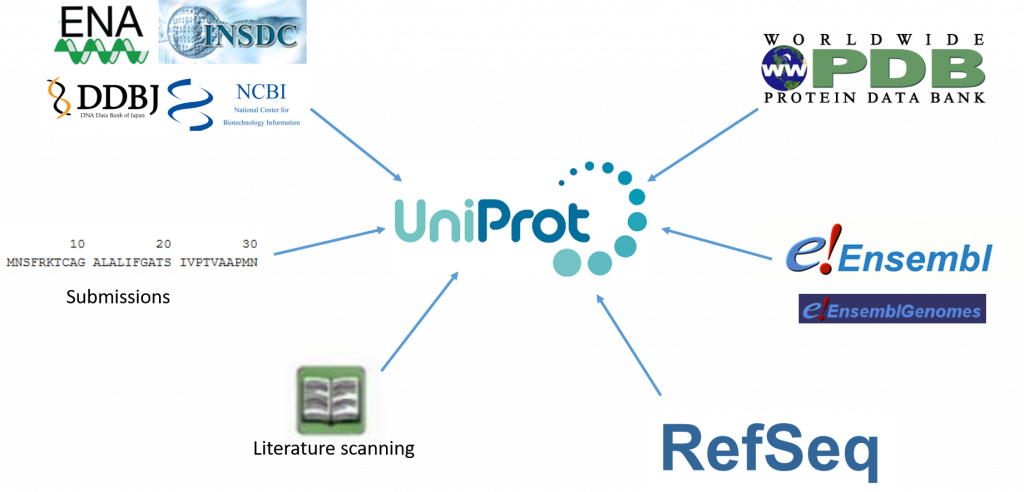
The UniProt knowledgebase is a large resource of protein sequences and associated detailed annotation. The database contains over 60 million sequences, of which over half a million sequences have been curated by experts who critically review experimental and predicted data for each protein. The remainder are automatically annotated based on rule systems that rely on the expert curated knowledge. Since our last update in , we have more than doubled the number of reference proteomes to , giving a greater coverage of taxonomic diversity. We implemented a pipeline to remove redundant highly similar proteomes that were causing excessive redundancy in UniProt.
The aim of the UniProt Knowledgebase is to provide users with a comprehensive, high-quality and freely accessible set of protein sequences annotated with functional information. In this publication we describe enhancements made to our data processing pipeline and to our website to adapt to an ever-increasing information content. The number of sequences in UniProtKB has risen to over million and we are working towards including a reference proteome for each taxonomic group. We continue to extract detailed annotations from the literature to update or create reviewed entries, while unreviewed entries are supplemented with annotations provided by automated systems using a variety of machine-learning techniques. In addition, the scientific community continues their contributions of publications and annotations to UniProt entries of their interest. The UniProt databases enable the research community to explore the diversity of life as described by the complement of proteins expressed by each organism. The UniRef databases cluster sequence sets at various levels of sequence identity and the UniProt Archive UniParc delivers a complete set of known unique sequences, including historical obsolete sequences. Data from selected resources are additionally integrated into UniProtKB records to add biological knowledge and associated metadata enabling the database to act as a central hub from which users can link out to other resources. Community functional annotation adds further value to the entry annotations. The integration of these data and the manual curation of protein features, such as functional domains and active sites, amino acid variants, ligand binding sites and post-translational modifications PTMs in the UniProt record, provide our users with mechanistic insights into how, for example, specific variants can lead to disease or resistance to a drug or to a pathogen.
Uniprot
All materials are free cultural works licensed under a Creative Commons Attribution 4. UniProt provides the scientific community with a comprehensive, high-quality and freely accessible resource of protein sequence and functional information. As the number of completely sequenced genomes continues to increase, huge efforts are being made in the research community to understand as much as possible about the proteins encoded by these genomes. This work is critical to many areas of science including biology, medicine and biotechnology — and is generating a wealth of data. UniProt provides an up-to-date, comprehensive body of protein information. The resource facilitates scientific discovery by collecting, interpreting and organising this information, which saves researchers countless hours of work. You can use UniProt for a wide range of tasks, from finding out about your protein of interest and comparing its protein sequence with other proteins, to mapping a list of identifiers from an external database to UniProtKB or vice versa. Around 90 people are involved across the three groups through a range of tasks such as database curation, software development and user support.
Tumblr original post finder
Abstract The aim of the UniProt Knowledgebase is to provide users with a comprehensive, high-quality and freely accessible set of protein sequences annotated with functional information. We are adapting our data input pipeline to ensure that we present a reference proteome for each taxonomic grouping to the research community. Revision received:. Population genetics. Finding enzyme cofactors in protein data bank. Citing articles via Web of Science Updated datasets from clinically relevant sources of sequence variation e. We have recently extended this approach to fungal species where there has been a similar if not as large as yet explosion in sequencing of highly similar genomes. This view is also now used when users search for specific information on one or a group of proteins, for example for proteins referenced in a specific citation or those assigned to a specific subcellular location, as it provides immediate access to the supporting data. Using WormBase ParaSite: an integrated platform for exploring helminth genomic data. The results of these methods are combined and refined according to UniProt standards with the addition of the appropriate UniProtKB annotation. The semi-automated rule-based computational annotation UniRule system 27 combines the detailed annotation found in the reviewed records in UniProtKB with the information on protein families predicted by InterPro, in order to create rules for propagating annotation to the unreviewed proteins in the database.
The aim of the UniProt Knowledgebase is to provide users with a comprehensive, high-quality and freely accessible set of protein sequences annotated with functional information. In this article, we describe significant updates that we have made over the last two years to the resource.
Computationally selected reference proteomes are chosen based on a number of criteria, including the level of curation reviewed versus unreviewed , protein name e. It is important to realize that there may be additional records for a particular species which do not belong to the defined proteome. Select Format Select format. Figure 6. UniProt Consortium Watkins X. Yates A. Exploring the dark genome: implications for precision medicine. Drysdale R. I agree to the terms and conditions. Consortium for Top Down Proteomics Proteoform: a single term describing protein complexity. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Google Scholar.

0 thoughts on “Uniprot”